Duck plague virus (DPV), belonging to herpesviruses, is a linear double-stranded DNA virus. Duck Plague (DP) is one of the most serious diseases in China's waterfowl industry, which often causes huge economic losses. Recently, increasing reports revealed that multiple long non-coding RNAs (lncRNAs) can possess great potential in the regulation of host antiviral immune response. Here, lncRNAs and mRNAs in DPV infected duck embryonic fibroblast (DEF) cells were identified by high-throughput RNA-sequencing (RNA-seq). In our study, 218 DE lncRNAs and 2840 DE mRNAs were obtained in DEF after DPV infection at 30 h post-infection (hpi). And we formed a complex regulatory network depending on in-silico analysis and prediction. Among these DEGs and target genes, some have been authenticated as immune-related molecules, such as a Macrophage mannose receptor (MR), Anas platyrhynchos toll-like receptor 2 (TLR2), leukocyte differentiation antigen, interleukin family, and their related regulatory factors. Furthermore, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis, we found that the target genes may have important effects on biological development, biosynthesis, signal transduction, cell biological regulation, and cell process. Also, we obtained, the potential targeting relationship existing in DEF cells between host lncRNAs and DPV-encoded miRNAs by software.
This study was published in BMC Genomics (IF=4.547), and titled as “Differential expression profile and in-silico functional analysis of long noncoding RNA and mRNA in duck embryo fibroblasts infected with duck plague virus”. Doi: 10.1186/s12864-022-08739-7.
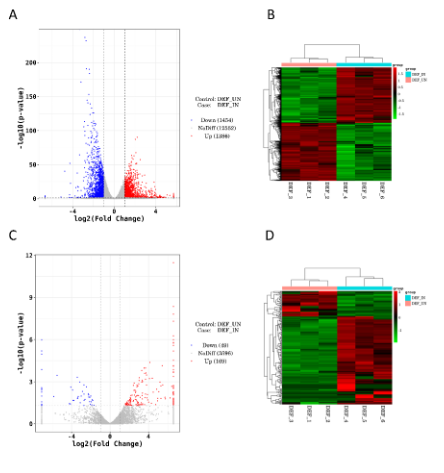
Fig.1 Differential analysis of lncRNA/mRNA expression in DEFs infected with DPV.

