Salmonella is a widespread foodborne pathogen with more than 2600 serotypes and concerning antimicrobial resistance (AMR). Waterfowl are an important source of Salmonella, but studies on the prevalence and resistance of Salmonella in waterfowl are still scarce. In this study, a total of 126 Salmonella isolates (65 from 2018-2020 and 61 from 2002-2005) were obtained from waterfowl samples. We systematically examined these isolates for their serotypes, pulsed-field gel electrophoresis (PFGE) types, and AMR phenotypes and genotypes. These isolates distributed in six serogroups and exhibited a diversity of PFGE characteristics, and all isolates were multi-drug resistant. Whole-genome sequencing of 49 isolates revealed that these isolates carried a wide variety of AMR genes, including multiple efflux pump genes and specific resistance genes. Interestingly, tet (A)/tet (B) and catII resistance genes were only detected in the earlier isolates, whereas gyrA (S83F, D87N, and D87G) and gyrB (E466D) mutations were more common in recent isolates, consistent with isolates from different periods having different resistance patterns. In addition, we detected eight plasmid replicons, suggesting that multiple antimicrobial-resistant plasmids spread in Salmonella, which may contribute to the horizontal transfer of AMR genes and provide a competitive advantage to Salmonella. Related studies are presented as "Phenotypic and genotypic characterization of antimicrobial resistance profiles in Salmonella isolated from waterfowl in 2002 -2005 and 2018-2020 in Sichuan, China" published online in Frontiers in Microbiology (IF=6.064), https://doi.org/ 10.3389/fmicb.2022.987613.
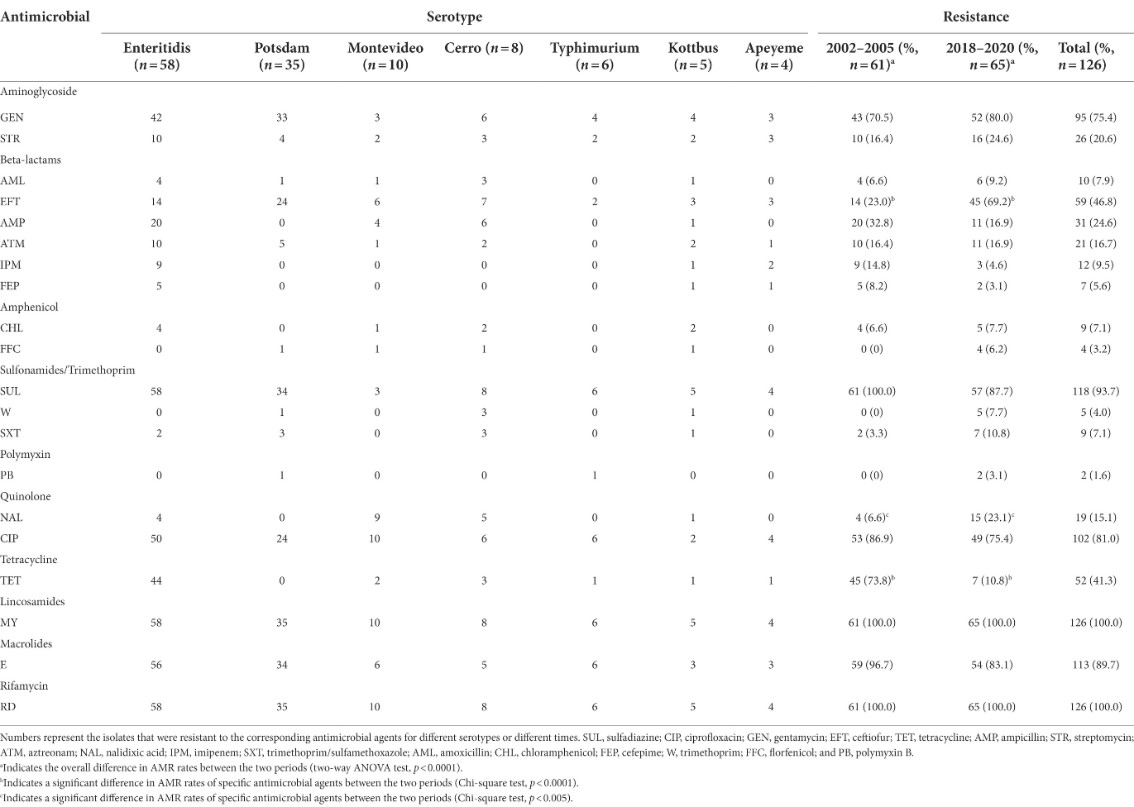
Drug resistance of Salmonella from waterfowl in Sichuan

